Home > Press > A needle in a haystack: How does a broken DNA molecule get repaired?
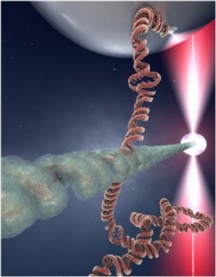 |
| After a DNA molecule breaks, the broken ends search for an intact DNA region with the same sequence in order to get repaired. The image shows an artist impression of the contact point between a RecA-protein DNA molecule (the "broken end"; horizontal) and a DNA molecule (vertical), where it is probed whether both molecules have the same sequence. If they do not, they will break the contact. If the same sequence is found however, the molecules stably bind and the repair process is initiated. The present study discovered the mechanism of the recognition process from dual molecule experiments where individual DNA molecules can be manipulated with beads.
Credit: Image courtesy Cees Dekker lab TU Delft / Tremani |
Abstract:
Scientists from the Kavli Institute of Nanoscience at Delft University of Technology have discovered a key element in the mechanism of DNA repair. When the DNA double helix breaks, the broken end goes searching for the similar sequence and uses that as a template for repair. Using a smart new dual-molecule technique, the Delft group has now found out how the DNA molecule is able to perform this search and recognition process in such an efficient way. This week, the researchers report their findings in Molecular Cell.
A needle in a haystack: How does a broken DNA molecule get repaired?
The Netherlands | Posted on May 3rd, 2012A staggering problem
Sometimes, the DNA double helix gets broken: both strands are accidentally cut. This presents a vital problem because cells cannot cope with such damaged DNA. Genomic DNA instabilities such as these, are a known cause of cancer. The good news is that an intricate DNA repair system exists which is impressively error-proof and efficient. How does this work?
First, proteins form a filamentous structure on the broken DNA end. Second, this filament examines recently copied DNA or the second DNA chromosome (remember that we have two copies of each chromosome) in search of a DNA sequence that matches that of the broken end. Note that this is a daunting task: given that, for example, our human genome contains three billion base pairs, finding your few hundred base pairs of interest, is really like finding a needle in a haystack.
'Still this search process occurs within minutes and with great efficiency. How that is achieved, has been a mystery for decades. The new experiments from our group now resolve this by revealing the key step in the process, the molecular recognition step', says scientist Iwijn de Vlaminck, who was the postdoc that did the experiments in the group of prof. Cees Dekker at Delft.
Search operation
'In bacteria, the so-called RecA protein is responsible for conducting the search operation. In E. coli bacteria, a filament of RecA protein formed on DNA, searches and pairs a sequence within a second DNA molecule with remarkable speed and fidelity. To do so, individual molecules of RecA first come together to form a filamentous structure on the broken DNA. The filament then grabs DNA molecules in its vicinity and compares their sequence to the sequence of the broken DNA. When a sequence match is found, both molecules bind tightly to one another allowing repair to ensue', says De Vlaminck (since recently at Stanford University).
'We found that the filament's secondary DNA-binding site interacts with a single strand of the incoming double-stranded DNA during homology sampling. Recognition is achieved upon binding of both strands of the incoming DNA to each of two DNA-binding sites in the filament.'
The data indicate that the fidelity of the search process is governed by the distance between the DNA binding sites. The Delft experiments clarify what exactly happens in the sequence comparison of the two molecules, making clear why a 'wrong' sequence leads to quick dissociation of the molecules while a 'correct' sequence makes a strong bond leading up to further repair. These are the two elements that lead to the impressive speed and high efficiency of the DNA repair process.
New instrument
The team from TU Delft developed a unique new instrument that makes it possible to independently manipulate an individual DNA molecule and an individual RecA filament and to measure the strength of intermolecular interactions. This dual-molecule manipulation instrument combines magnetic-tweezer and laser-trapping-based DNA-molecule manipulation with a laminar flow system. The setup also allowed to unwind the DNA helix a bit, thus opening local regions where the normal duplex was destabilized. This effect turned out to be crucial for the molecular recognition process. Thus, the team was able to directly probe the strength of the two-molecule interactions involved in search and recognition and build a new model for it.
####
For more information, please click here
Contacts:
Ilona van den Brink
31-152-784-259
Copyright © Delft University of Technology
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related News Press |
News and information
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Nanomedicine
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Discoveries
![]() From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Announcements
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||








