Home > Press > nanoCAGE reveals transcriptional landscape of the mouse main olfactory epithelium
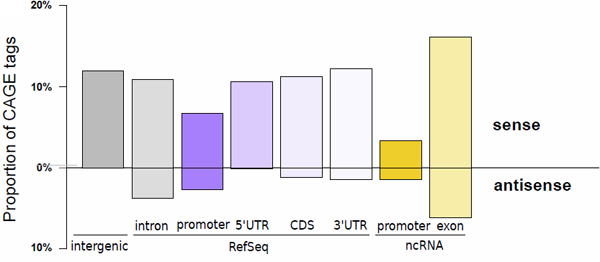 |
| Figure 1 MOE transcription start sites recapitulate known transcript initiation and reveal the extent of non-coding transcripts. Histogram depicting the proportion of tags aligned to the proximal promoter of transcript models (defined as the region spanning from the 5' end to 500 bp upstream), the 5' UTR, the coding sequence (CDS), the 3' UTR (in decreasing purple colors), the proximal promoter of FANTOM3 non-coding RNA (in orange), and the FANTOM3 non-coding RNA (in light orange). The upper part of the bar plot shows TSSs located on the same strand as the annotation, while the lower part depicts TSSs located on the opposite strand. The percentage of TSSs that do not colocalize with any of those annotations is represented by the grey bar. |
Abstract:
The problem in biology of how to identify the promoters of olfactory receptor genes (>1000 genes) has remained unsolved due to the difficulty of purifying sufficient material from the olfactory epithelium. Researchers at the RIKEN Omics Science Center, collaborating with scientists from Italy, Norway, the United States, the United Kingdom and Germany, have now solved this problem using nanoCAGE technology, which enables comprehensive analysis of transcription start sites (TSSs) from tiny biological samples.
nanoCAGE reveals transcriptional landscape of the mouse main olfactory epithelium
Japan | Posted on January 5th, 2012In mouse, odor is sensed by the main olfactory epithelium (MOE) by about 1100 types of olfactory receptors that are expressed by olfactory sensory neurons. Interestingly, each sensory neuron expresses only a single type of olfactory receptors, whose selective expression mechanism remains largely unknown. The population of olfactory sensory neurons that express a given olfactory receptor is small, which makes transcription analysis difficult.
Researchers at the RIKEN Omics Science Center recently developed nanoCAGE (CAGE: Cap Analysis of Gene Expression), the only technology that can comprehensively identify precise TSSs of both protein-coding and non-coding capped mRNAs and quantify their individual levels of expression starting from tiny biological samples of only a few nanograms of RNA (Plessy et al., Nature Methods, 7, 528-534, 2010). By using nanoCAGE on the MOE, the researchers succeeded in identifying 87.5% of the olfactory receptor gene TSSs. The results show for the first time that olfactory receptor genes contain hundreds of non-coding RNAs, suggesting that these RNAs may play important roles in the transcriptional regulation of olfactory receptors.
Dr. Piero Carninci commented, "Combined with CAGE, nanoCAGE technology provides a new opportunity to unveil gene networks in the nervous system using omics approaches."
The research is published in the journal Genome Research.
Reference:
Plessy C, Pascarella G, Bertin N, Akalin A, Carrieri C, Vassalli A, Lazarevic D, Severin J, Vlachouli C, Simone R, Faulkner GJ, Kawai J, Daub CO, Succhelli S, Hayashizaki Y, Mombaerts P, Lenhard B, Gustincich S, Carninci P. "Promoter architecture of mouse olfactory receptor genes." Genome Research, 2011, DOI: 10.1101/gr.126201.111
####
About Riken Research
RIKEN is Japan's flagship research institute devoted to basic and applied research. Over 2500 papers by RIKEN researchers are published every year in reputable scientific and technical journals, covering topics ranging across a broad spectrum of disciplines including physics, chemistry, biology, medical science and engineering. RIKEN's advanced research environment and strong emphasis on interdisciplinary collaboration has earned itself an unparalleled reputation for scientific excellence in Japan and around the world.
About the Omics Science Center
Omics is the comprehensive study of molecules in living organisms. The complete sequencing of genomes (the complete set of genes in an organism) has enabled rapid developments in the collection and analysis of various types of comprehensive molecular data such as transcriptomes (the complete set of gene expression data) and proteomes (the complete set of intracellular proteins). Fundamental omics research aims to link these omics data to molecular networks and pathways in order to advance the understanding of biological phenomena as systems at the molecular level.
Here at the RIKEN Omics Science Center, we are developing a versatile analysis system, called the "Life Science Accelerator (LSA)", with the objective of advancing omics research. LSA is a multi-purpose, large-scale analysis system that rapidly analyzes molecular networks. It collects various genome-wide data at high throughput from cells and other biological materials, comprehensively analyzes experimental data, and thereby aims to elucidate the molecular networks of the sample. The term "accelerator" was chosen to emphasize the strong supporting role that this system will play in supporting and accelerating life science research worldwide.
For more information, please click here
Contacts:
Piero Carninci
Functional Genomics Technology Team
RIKEN Omics Science Center
Tel: +81-(0)45-503-9222 / Fax: +81-(0)45-503-9216
Yokohama Planning Section
RIKEN Yokohama Research Promotion Division
Tel: +81-(0)45-503-9117 / Fax: +81-(0)45-503-9113
Global Relations Office
RIKEN
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Copyright © Riken Research
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related News Press |
News and information
![]() Simulating magnetization in a Heisenberg quantum spin chain April 5th, 2024
Simulating magnetization in a Heisenberg quantum spin chain April 5th, 2024
![]() NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
![]() Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Discoveries
![]() Chemical reactions can scramble quantum information as well as black holes April 5th, 2024
Chemical reactions can scramble quantum information as well as black holes April 5th, 2024
![]() New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
![]() Utilizing palladium for addressing contact issues of buried oxide thin film transistors April 5th, 2024
Utilizing palladium for addressing contact issues of buried oxide thin film transistors April 5th, 2024
Announcements
![]() NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
![]() Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Nanobiotechnology
![]() New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
![]() Good as gold - improving infectious disease testing with gold nanoparticles April 5th, 2024
Good as gold - improving infectious disease testing with gold nanoparticles April 5th, 2024
![]() Researchers develop artificial building blocks of life March 8th, 2024
Researchers develop artificial building blocks of life March 8th, 2024
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||








