Home > Press > Cells traverse developmental divide via Blimp
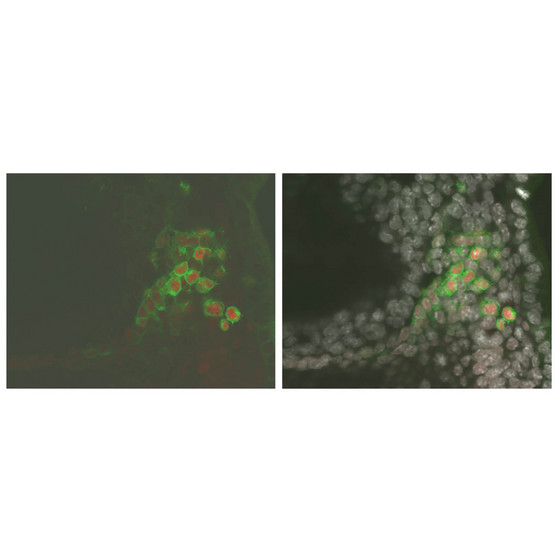 |
| Figure 1: Sox2 is among the genes upregulated by Blimp1 as part of the PGC developmental program. These cells have been fluorescently labeled to illustrate this process; Blimp1-expressing cells are labeled green, Sox2 protein is labeled red. In the right panel, cell nuclei have also been labeled with a generic staining agent and pseudo-colored white to reveal all individual cells in the specimen. Shown is a posterior part of a day 7.5 embryo. |
Abstract:
A method for single-cell genomic profiling has helped researchers to identify a putative ‘master switch' for reproductive cell development in the mouse embryo
Cells traverse developmental divide via Blimp
Japan | Posted on October 31st, 2008An animal's reproductive capabilities are established early in development, when a homogeneous embryonic cell population gives rise to two distinct cell types—somatic cells that form the vast majority of body tissues, and primordial germ cells (PGCs) that ultimately yield spermatozoa or ova.
Identifying genes responsible for ‘programming' PGC development will be essential to fully understand this essential developmental process. Unfortunately, existing techniques for large-scale gene expression profiling are designed for use with multicellular samples—an ineffective strategy for PGC analysis.
"PGCs are small in number—especially at early stages—and are embedded in somatic neighbors," explains Mitinori Saitou, of the RIKEN Center for Developmental Biology in Kobe. "Therefore, for systematically identifying genes specific to PGCs, single-cell analysis is considered to be essential." Prior work from Saitou's team identified several genes potentially important to PGC development. Now, his group has developed a powerful new technique for preparation and amplification of nucleic acids from individual cells, enabling stage-specific genomic profiling of mouse PGCs in unprecedented detail1.
The researchers focused on identifying genes regulated by Blimp1, a gene identified in their earlier work2. After analyzing PGCs from various developmental stages, it became clear that Blimp1 expression specifically increases in these cells over time. They also observed that although early-stage PGCs exhibit expression profiles for certain developmental genes that are similar to those observed in somatic cells, continued expression of Blimp1 leads to reversal of these expression patterns, actively driving development onto a PGC-specific trajectory (Fig. 1).
A broader comparison of stage-specific gene expression in PGCs and somatic cells enabled Saitou's team to assemble clusters of genes that are generally up- or down-regulated by Blimp1, allowing them to be categorized respectively as ‘specification' or ‘somatic' genes. Certain gene types were enriched for each category—including cell division regulators for the somatic genes and effectors of germ cell development for the specification genes—and each category also contained distinct sets of genes involved in embryonic development and body pattern formation.
Follow-up analyses confirmed that Blimp1 plays a central role in managing appropriate regulation of both somatic and specification genes for PGC development. "To me, the fact that Blimp1 represses essentially all the genes normally repressed in PGCs in comparison to their somatic neighbors is the most important finding," says Saitou. Now, having glimpsed the ‘big picture', Saitou's team hunting for the primary target genes for Blimp1, and the mechanism by which it switches them on to set PGC development in motion.
Reference
1. Kurimoto, K., Yabuta, Y., Ohinata, Y., Shigeta, M., Yamanaka, K. & Saitou, M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes & Development 22, 1617-1635 (2008).
2. Ohinata, Y., Payer, B., O'Carroll, D., Ancelin, K., Ono, Y., Sano, M., Barton, S.C., Obukhanych, T., Nussenzweig, M., Tarakhovsky, A., et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207-213 (2005).
The corresponding author for this highlight is based at the RIKEN Laboratory for Mammalian Germ Cell Biology
####
For more information, please click here
Copyright © Riken
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related Links |
| Related News Press |
News and information
![]() Simulating magnetization in a Heisenberg quantum spin chain April 5th, 2024
Simulating magnetization in a Heisenberg quantum spin chain April 5th, 2024
![]() NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
![]() Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Nanomedicine
![]() New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
![]() Good as gold - improving infectious disease testing with gold nanoparticles April 5th, 2024
Good as gold - improving infectious disease testing with gold nanoparticles April 5th, 2024
![]() Researchers develop artificial building blocks of life March 8th, 2024
Researchers develop artificial building blocks of life March 8th, 2024
Discoveries
![]() Chemical reactions can scramble quantum information as well as black holes April 5th, 2024
Chemical reactions can scramble quantum information as well as black holes April 5th, 2024
![]() New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
![]() Utilizing palladium for addressing contact issues of buried oxide thin film transistors April 5th, 2024
Utilizing palladium for addressing contact issues of buried oxide thin film transistors April 5th, 2024
Announcements
![]() NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
![]() Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||








