Home > Press > Nanoscale tool allows scientists to study membrane proteins one at a time
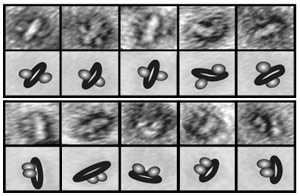 |
| Isolation chamber. A new tool developed at Rockefeller allows scientists to study membrane proteins individually, or in pairs, to see how they interact with other molecules. The scientists use an electron microscope to take images of isolated NABBs and categorize the orientation of the receptors they contain as either antiparallel (top) or parallel (bottom). |
Abstract:
In biology, as in construction, it's all about having tools that fit the job. Researchers at Rockefeller University have now created a tiny tool, more than 10,000 times smaller than the diameter of a human hair, capable of encasing single membrane proteins from living cells. The new system, which resembles a nanoscale sushi roll, will allow investigators to individually stimulate these key proteins with specific molecules and signals in order to precisely define the biological reactions that result.
Nanoscale tool allows scientists to study membrane proteins one at a time
New York, NY | Posted on March 7th, 2008The Nanoscale Apolipoprotein Bound Bilayers (NABBs), developed by scientists in Rockefeller's Laboratory of Molecular Biology and Biochemistry and reported in the Journal of Molecular Biology, is a versatile device that can likely be adapted to any of the myriad transmembrane receptors that direct cell activity by reacting to molecules outside the cell and activating signals inside the cell.
"Today it is impossible to know exactly what a single protein on the surface of a cell that has thousands of other proteins is doing. It might be acting on its own or binding to one or more other proteins," says Thomas Sakmar, Richard M. and Isabel P. Furlaud Professor and head of the laboratory and the study's senior investigator. "With this tool, we can control the receptor's membrane environment and test all possibilities of interaction with ligands, other receptors or other proteins. It's one way to figure out how a complex system works."
Previously, researchers studied the functions of such proteins by investigating literally millions of them floating together in a soup created when cell membranes are broken apart and solubilized chemically. But this method of studying proteins is problematic, the researchers say: The membrane protein mixtures tend to be inhomogeneous and it is difficult — partially due to poor stability of the isolated proteins — to purify them in their active state in order to understand what the receptors are doing individually.
The solution, devised by Sakmar, first author Sourabh Banerjee, a graduate student in the Tri-Institutional Chemical Biology Program, and Thomas Huber, a postdoc, arose as the team searched for a way to exquisitely catalogue the functions of individual G-protein-coupled receptors (GPCRs), a large family of transmembrane proteins that are involved in many diseases and are often the target of medicinal agents. The structure they built was developed using a hard-working human transport particle, the high-density lipoprotein (HDL), as a model system. This flat, circular structure is essentially a complex of phospholipids belted together by apolipoprotein A-I (apo A-I) to carry cholesterol and lipids through blood to the liver.
Assuming that evolutionary forces might have already optimized a biological solution to an engineering problem, Huber suggested using apo A-I from zebrafish. "Based on the sequence of zebrafish apo A-I, we thought that it may yield structurally homogeneous discs," Banerjee says. So in their NABB, zebrafish apo A-I (known as zap1) forms a belt that makes two layers of lipids stick together — like the seaweed that keeps sticky rice together in sushi.
They then devised a method to trigger rapid self-assembly of these disc-like nanoparticles from mixtures of zap1, lipids and extracted cellular membrane proteins. "We have made it fairly straightforward to make these structures and they form in less than an hour," says Banerjee, who coined the term NABBs.
The team visualized individual antibody fragments bound to the receptors with an electron microscope. And, as a proof of principle, they experimented with rhodopsin, a prototypical GPCR. They found that rhodopsin was remarkably stable in NABBs — as stable as in its native membranes. They also found that it doesn't require a "dimer," or union of two rhodopsin receptors, to produce a response — as many scientists had argued — but that rhodopsin can be activated in its monomeric form.
"Each protein is very happy inside its own disc and the beauty is that both sides of these receptors, the part that is inside the cell and the part that is outside, are exposed to whatever you want to test it with," Sakmar says. "That way we can use it to monitor what happens on both sides of the cell membrane."
"This tool can be used for a wide variety of membrane proteins," Banerjee says. "We think it will be important for high-throughput screening for new drugs that can bind to membrane proteins involved in disease."
Journal of Molecular Biology online: February 2, 2008
####
About Rockefeller University
The university’s laboratory-based organizational structure “without walls” and pared-down layers of administration do away with the schools and academic departments that too often separate scientists. “This approach fosters a tremendously rich soup of interdisciplinary research and collaboration,” says Rockefeller professor and Nobel laureate Günter Blobel.
For more information, please click here
Contacts:
Joseph Bonner
212-327-8998
Copyright © Rockefeller University
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related News Press |
News and information
![]() Simulating magnetization in a Heisenberg quantum spin chain April 5th, 2024
Simulating magnetization in a Heisenberg quantum spin chain April 5th, 2024
![]() NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
![]() Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Nanomedicine
![]() New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
![]() Good as gold - improving infectious disease testing with gold nanoparticles April 5th, 2024
Good as gold - improving infectious disease testing with gold nanoparticles April 5th, 2024
![]() Researchers develop artificial building blocks of life March 8th, 2024
Researchers develop artificial building blocks of life March 8th, 2024
Discoveries
![]() Chemical reactions can scramble quantum information as well as black holes April 5th, 2024
Chemical reactions can scramble quantum information as well as black holes April 5th, 2024
![]() New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
![]() Utilizing palladium for addressing contact issues of buried oxide thin film transistors April 5th, 2024
Utilizing palladium for addressing contact issues of buried oxide thin film transistors April 5th, 2024
Announcements
![]() NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
NRL charters Navy’s quantum inertial navigation path to reduce drift April 5th, 2024
![]() Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Discovery points path to flash-like memory for storing qubits: Rice find could hasten development of nonvolatile quantum memory April 5th, 2024
Tools
![]() Ferroelectrically modulate the Fermi level of graphene oxide to enhance SERS response November 3rd, 2023
Ferroelectrically modulate the Fermi level of graphene oxide to enhance SERS response November 3rd, 2023
![]() The USTC realizes In situ electron paramagnetic resonance spectroscopy using single nanodiamond sensors November 3rd, 2023
The USTC realizes In situ electron paramagnetic resonance spectroscopy using single nanodiamond sensors November 3rd, 2023
Nanobiotechnology
![]() New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
New micromaterial releases nanoparticles that selectively destroy cancer cells April 5th, 2024
![]() Good as gold - improving infectious disease testing with gold nanoparticles April 5th, 2024
Good as gold - improving infectious disease testing with gold nanoparticles April 5th, 2024
![]() Researchers develop artificial building blocks of life March 8th, 2024
Researchers develop artificial building blocks of life March 8th, 2024
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||








